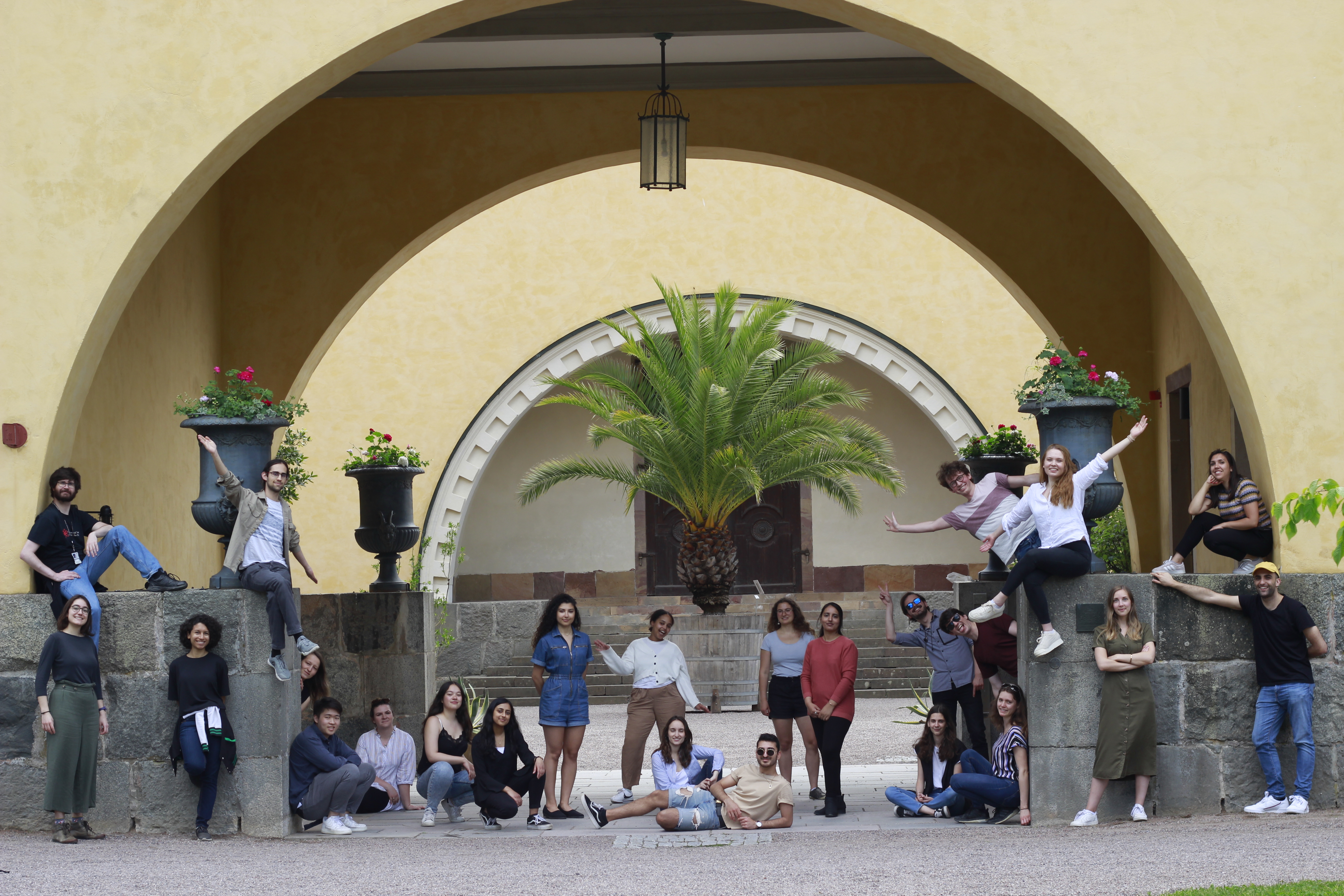iGEM Uppsala 2021
Who are we?
The Uppsala iGEM 2021 team consists of 20 motivated students from all over the world studying at 8 different programmes at Uppsala University.
What is the project?
This years project will be within the field of cultivated meat which is at a fast paced development and we’re seeking to solve a problem that is holding back it’s growth due to the high standards for cultivation and their cost
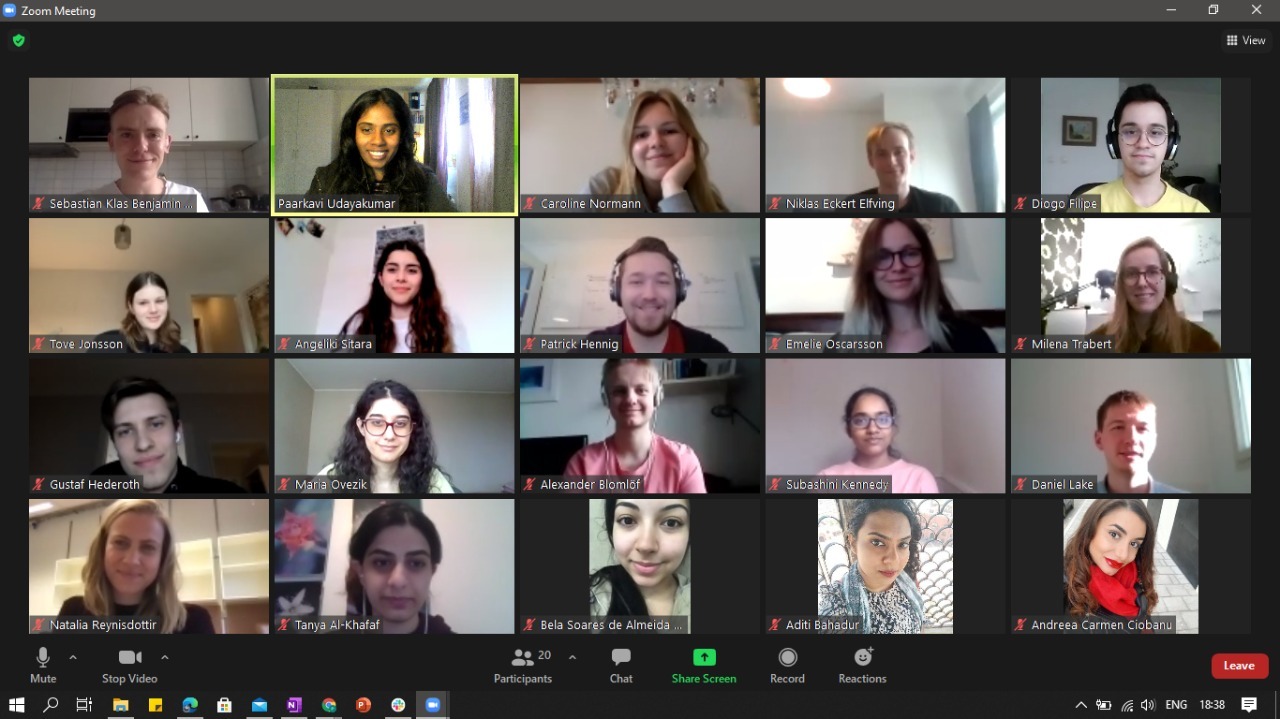
iGEM Uppsala 2020
NANOFLEX
Who are we?
The Uppsala 2020 team consists of 25 brilliant students from different faculties at Uppsala University and from across the globe. From engineers in biotechnology, computational scientists and biologists all are united in the iGEM Uppsala spirit.
What is the project?
This year's project, NANOFLEX, is an ambitious one. Team has created a modular cellular biosensor. By using exchangeable nanobodies fused to a DNA binding domain, NANOFLEX is able to detect a target molecule and induce an output signal visible by the naked eye. A key part of the project is the modular approach; this allows the technology to be applied in a variety of situations ranging from environmental detection to drug detection. Furthermore, the team has laid the groundwork of a more efficient assembly, Type IIS iGEM standard, for the many genetic elements of NANOFLEX. The team is also working on making the biosensor portable, further increasing its utility.
Check out the 2020 project presentation in the video below!
Want to know more?
Feel free to visit the team wiki! If you have any questions you are welcome to contact the team via the following email address:
igemuppsala2020@gmail.com
Visit the team wiki
View moreiGEM Uppsala 2019
Biomass Destruction
Why Lignin?
In the global, highly technological world of the 21st century, it is easy to forget the immense scientific and engineering challenges that have been overcome to produce the everyday products that many have come to expect and rely upon. When we print out a document, read the newspaper or open our textbooks, we are enjoying a deceptively simple commodity: paper. In reality, paper is the end result of a lengthy and multi-step process called wood pulping. In wood pulping there is approximately 7 tonnes of black liquor which is a highly viscous aqueous waste. This is produced during the alkaline Kraft process of manufacturing one tonne of pulp. A specific heterogeneous polymer, lignin, makes up around 45% in the solid composition of Black liquor, leading to the production of numerous tons of lignin every year.
Currently, over 90% of lignin is burned to recover the chemicals from black liquor. This can be considered massive waste due to the potential for creating enormous economicvalue from the lignin in black liquor. For example lignin can be used as a raw material for the production of vanillin, phenols.
Our aim
In this project we plan to use enzymes related to wood decay, originating from the most effective bio-degraders of wood: fungi. White-rot fungi are the most effective bi-degraders of lignocellulosic biomass. The Ligninolytic enzymatic consortium produced by white-rot fungi is mostly composed by laccases, peroxidases and H2O2 supplying enzymes.
Who are we?
We are a team of enthusiastic science explorers coming from different academic backgrounds, from biology, chemistry, healthcare, and molecular engineering. We represent the spirit of a university: different cultures, tongues, and countries from the entire globe: Swedish, English, Hindi and Spanish. From the warm shores of south america and southeast asia to cold wind and waters of the baltic in northern europe. We are the iGEM Uppsala team!
Contact Information
If you have any questions about this project or want to contact the team, please email the project managers at:
projectleader@igemuppsala.se.

Visit their wiki
View moreiGEM Uppsala 2018
Worm Busters!
Our Project
All over the world horses are treated with large amounts of anthelmintics with the aim of killing parasites, regardless of need. These parasites are not deadly in small amounts, but when a critical mass is reached they can cause immense problems for domestic ungulates, including death. Since diagnostic methods for worm detection are not sufficiently advanced to properly gauge when animals should be treated, anthelmintic drugs are often administered at times when there’s no need to do so. This common practice is rapidly contributing to anthelmintic resistance of various nematode species. Currently the only method for detection of these are counting stool samples, which is both inaccurate and expensive. In order to address this issue, we are developing a smart bacteria that can detect and report the presence of common blood worms.

How will we achieve this?
We have a two pronged plan for our project. One is to engineer E. coli to give off a detectable signal when they’re near a worm, and the other is to engineer these same E. coli to give off a signal that attracts worms towards them. We plan to try this method out for both large blood worms, which we would try to detect in grass samples, and small blood worms, which would be detected while still inside the horse. A simple test with our designed worm buster would allow farm animal owners to better decide when to medicate their animals, reducing anthelmintic usage, or to know if a certain field is infected or not, and therefore let their animals graze somewhere else.
While the project mainly deals with parasites in horses, it is also applicable towards nematodes that infect other farm animals like sheep as well.
Who are we? Meet the team!
We are a total of 23 very dedicated students at various stages in our education; a mix of bachelors and masters level students. Our team is composed of individuals from various programmes including biotechnology, chemical engineering, nanotechnology, and lastly IT and computer science
Contact Information
If you have any questions about this project or want to contact the team, please email the project managers at:
projectleader@igemuppsala.se.

Visit their wiki
View moreiGEM Uppsala 2017
Crafting Crocin

The Team
The iGEM Uppsala team of 2017 was the most multicultural team so far! Only half of the 20 members had swedish ethnicity. Together, they explored the biochemistry of saffron, both in the lab and in the kitchen!Project overview
In 2017, iGEM Uppsala was Crafting Crocin. Crocin is an apocarotenoid (organic pigment) found in saffron, extracted from the flower Crocus Sativus. Crocin, crocetin, zeaxanthin and other pigments in the crocin pathway, are the compounds which gives saffron it’s beautiful red color and delicious taste. The pathway compounds have great potential as organic dyes for industrial applications. On top of this, recent studies suggest that these colorful components have several medicinal properties. By using synthetic biology for recombinant expression in Escherichia Coli we could reduce the price of the compounds within the crocin pathway and open up the possibility for industrial and medicinal applications to be further explored.Results
The team successfully engineered and E. coli strain that could produce the yellow saffron pigment zeaxanthin, and created BioBricks of the enzymes needed to produce crocin. Our team were the first ever to purify and prove activity of one of these enzymes, CsADH. In Boston, this years project got awarded a gold medal and the prize Best Manufacturing Project.
Visit their wiki
View moreiGEM Uppsala 2016
Chipgineering - Microfluidics, now for everyone

The Team
The iGEM Uppsala 2016 team consisted of 18 students studying biotechnology, biology, biochemistry and chemistry.Project overview
2016 years’ project centered around simplifying usage of two new exciting technologies: CRISPR/Cpf1 and microfluidics as well as characterising a new fluorescent eukaryotic protein UnaG.During the summer, the team developed a method to make microfluidics more available to iGEM teams and researchers and managed to express and characterize the UnaG protein. The team also made several biobricks for expressing UnaG as well as Cpf1 and its associated genes.
Results
The microfluidic subproject focused mainly on the development of a heat shock transformation-capable chip and on creating a blueprint/guide for making other microfluidic chips. The efforts were met with success and the guide, blueprint files and heat shock results were uploaded to the wiki.UnaG is an eukaryotic green fluorescent protein produced in Unagi eels. UnaG is of special interest due to it being smaller than the conventional GFP, making it suitable as a fusion protein. Furthermore, it only fluoresces upon binding of its ligand, bilirubin. The UnaG subproject was successful with the creation of three biobricks related to it and proof of expression in E. coli.
The CRISPR system is an interesting new protein system discovered in bacteria used for cutting and modifying DNA. Cpf1 is a variant of this system which creates overhangs making the process milder on the bacteria and allowing for easier gene insertion. The team managed to create two biobricks related to the protein but encountered obstacles during the summer and could not confirm that the sequence was correct.
The team presented its project in the Hardware track, choosing to focus its presentation on the microfluidics subproject. The project of iGEM Uppsala 2016 subsequently reached the Gold Metal criterias and won the award for Best project in Hardware. The team also participated in NiC 2016 hosted by the Stockholm team.
This project was rewarded a gold medal and won the price for best project in the hardware track, overgrad!

Visit their wiki
View moreIGEM Uppsala 2015
Decyclifier - The one PAHthway to rule them all
 The team of Uppsala iGEM 2015
The team of Uppsala iGEM 2015The 2015 team was the most diverse team in Uppsala iGEM history so far. Students of biotechnological engineering, bioinformatics, biology, chemistry, biomedicine and pharmacology all came together to battle in a lab for the summer in order to construct some bacteria capable of changing the world for the better!
Project overview
Polycyclic aromatic hydrocarbons (PAHs) are potent carcinogens produced by various activities ranging from grilling meat to coal gasification. In 2015 Uppsala iGEM aimed to degrade high molecular weight PAHs present in industrial waste from district heating plants. There is currently no sustainable way to handle this waste and therefore it is deposited in landfills. The team wanted to solve this problem by employing a bacterial based system.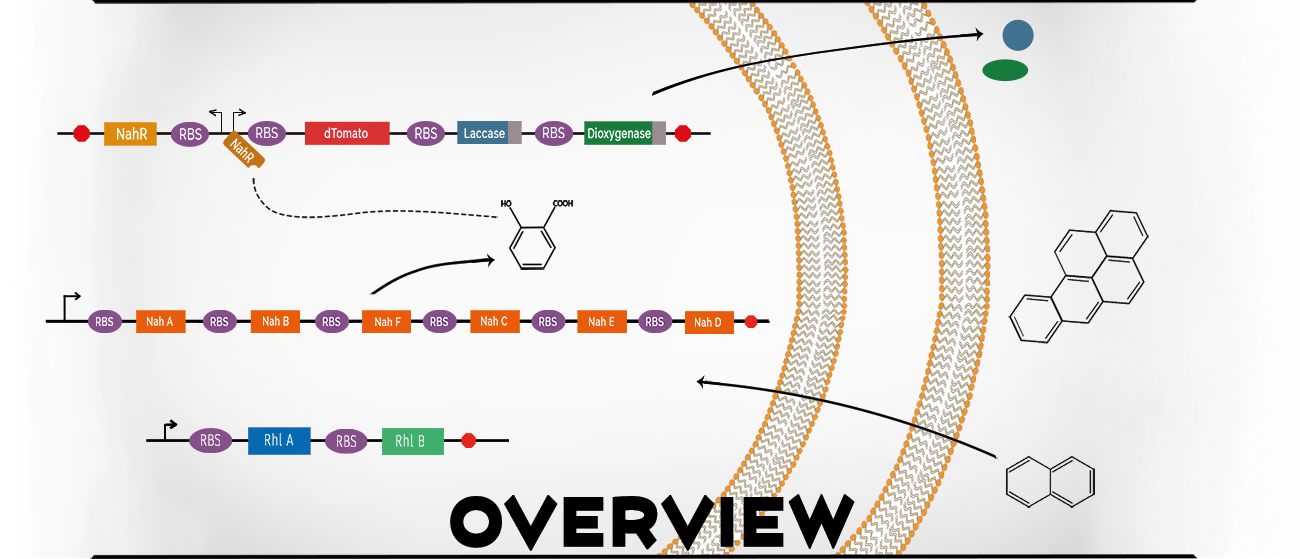
The team wanted their cells to be able to detect the PAHs for direct degradation. However, the high molecular weight PAHs do not readily pass through the cell membrane. They solved this obstacle by using the low weight PAH, naphthalene, as an indicator for the presence of heavier PAHs. The degradation of this small PAH inside the cell relieves repression of genes under the control of the NahR/Psal promoter system. This causes the expression of a series of enzymes for PAH degradation which are secreted outside the cell where they can oxidize and cleave the ring structures of the carcinogenic compounds. This in turn makes them available for downstream biodegradation. To increase the degradation efficiency, the system also produces rhamnolipids. These are biosurfactants that prevent the PAHs from aggregating, making them easier for the enzymes to access.
Results
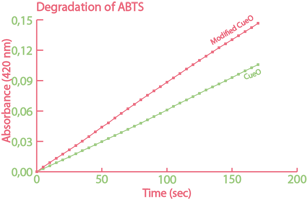
Some of the most important results from the project 2015 are summarized in the figures below. To read more about this project, to see how biosurfactants called rhamnolipids were expressed and also to read about how the team built their own, home made fluorometer, please visit their wiki.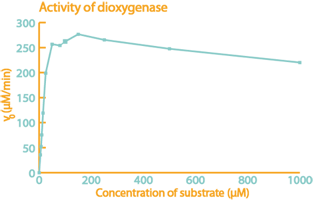 |
 |
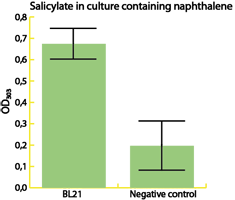 |
| The enzyme catechol-1,2-dioxygenase was characterized. | A modified laccase was shown to have higher activity than the wild-type. | Bacteria with our naphthalene-pathway were shown to break down naphthalene to salicylate. |
This project was rewarded gold medal and nominated for best environmental project, overgrad, at the Giant Jamboree in Boston.
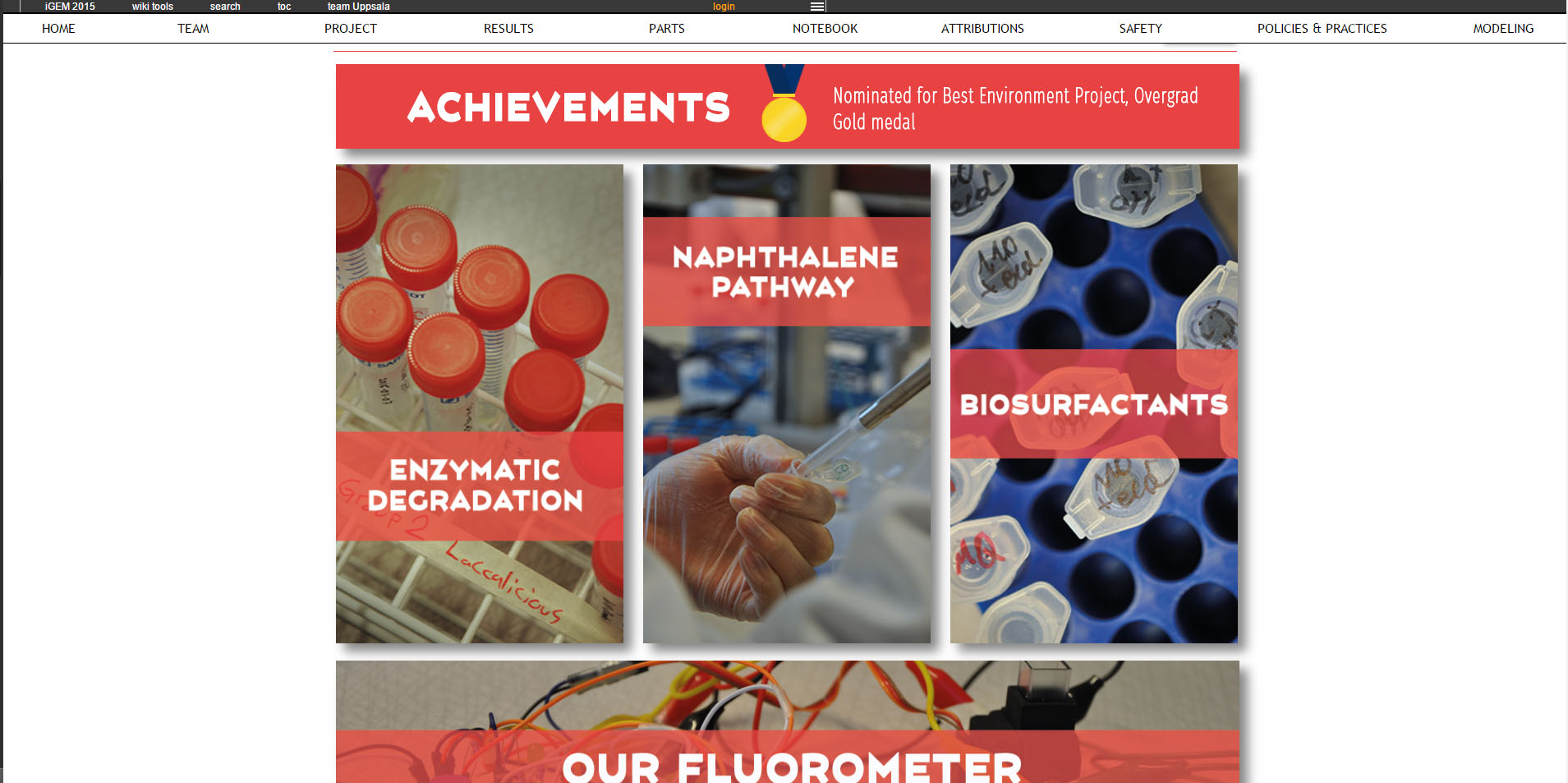
Visit their wiki
View moreIGEM Uppsala 2014
Bactissiles - The future of microbial combat
 The team of Uppsala iGEM 2014
The team of Uppsala iGEM 2014The project from the Uppsala team 2014 aimed to create a "missile" bacterium that would home towards a target and secrete an affectant to ensure a certain outcome. Such a system could have applications in a number of different fields, though the team chose to put this into practice in pinpointing pathogen-killing approach. In the prototype system, introduced into E. coli, the team hijacked the quorum sensing system of the gut pathogen Yersinia enterocolitica. This quorum sensing system, also called the Yen system, will activate/deactivate the targeting module and the killing module. The targeting module consists of a chemotaxis controlling element, while the killing module is made up of a bacteriocin, specific for Y. enterocolitica.
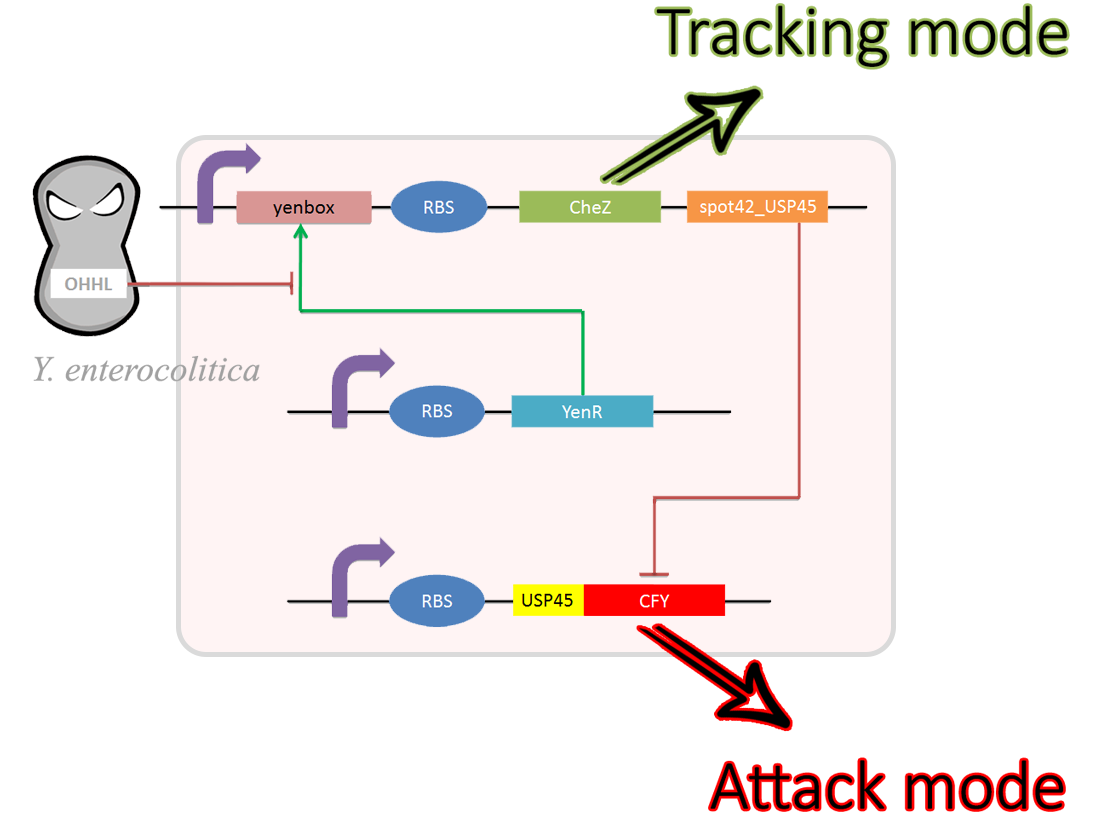
The regulatory circuit in the Bactissile
The bacteria, was given the commercial name the Bactissile. It was developed to be able to sense the presence of the pathogen, accumulate in its vicinity and emit a pathogen-specific bacterocin. By applying specificity in both the targeting mocule and the killing module, we can prevent unnecessary damage in the surrounding gut flora cultures.
Policy Practices
The policy practice was focused on creating a fictional company based on the project idea to see if it was viable as a commercial project. Apart from this they also continued with SynbioDay from previous year. The event was arranged in the Siegbahn lecture hall in Ångströms laboratory and was called the SynBioHour. During this event an introduction to synthetic biology was held, the team presented their project and also invited the team ULB_Brussel from Belgium to present their.
Results

The team managed to assemble and characterize each module separately. With the use of a flow cytometer they could show that the quorum sensing system, originally taken from Y. enterocolitica, coupled to a green fluorescence protein did actually work in E. coli and the amount of production could be regulated.
The targeting module could be shown to work using a knock-out E. coli strain where the chemotaxis controlling gene, CheZ, had been knock-out in the chromosome. By reintroducing CheZ into this strain it could be shown that the movement could be restored.
To characterize the killing module, the team had the privilege to go to the Swedish food authority Livsmedelsverket, to work with the actual pathogen. By culturing the pathogen Y. enterocolitica together with the bacterocin produced by the modified E. coli containing the killing module, it could be seen that the bacterocin did indeed kill Y. enterocolitica.
The team did unfortunately not have time to assemble all the modules together to the final system, but the results for the separate modules indicates that the final system should work as intended.
To hear their presentation, please watch this video.
When presenting the project at the Jamboree in Boston, they received very positive feedback and was also rewarded with a gold medal.
Visit their wiki
View moreIGEM Uppsala 2013
 The team of Uppsala iGEM 2013
The team of Uppsala iGEM 2013The goal of iGEM Uppsala 2013 project was to battle malnutrition with the help of a yoghurt. The project was divided into two main parts. building the parts for a recombinant lactobacillus and producing good and healthy compounds by metabolic engineering.
The first necessary problem to battle was to build the parts for the probiotic bacteria. The one we decided to use was the lactobacillus chassi that will be used to produce the nutritious compounds in the yoghurt. Lactobacillus is a probiotic bacteria that naturally exists in the gut flora and is thus seen as safe to eat.
The second part of the project was to produce the nutrients in the yoghurt, this was done with the help of metabolic engineering in bacteria. By introducing enzymes from several organisms, chosen compounds can be created. The compounds we tried to create was beta-carotene and a few other antioxidants in the carotenoid pathway. We also tried to create miraculin as well as the three main compounds of saffron to improve the taste of the yoghurt. Lastly we created p-Coumaric acid as well as resveratrol, which we had great success with.
Human Practice
The most important part of our human practice was to create and host Uppsala SynBioDay, a yearly event that invites the public to come and meet companies as well as the academic world and to understand what biotechnology and synthetic biology is.
Result
In the regional championship in France, Lyon we managed to get a Gold medal for our work as well as winning the prize for best natural biobrick. The prize we got was for our great characterization of the enzyme Tyrosine ammonia lyase producing the substance p-Coumaric acid from tyrosine. Thanks to these achievements we managed to proceeded to the final at MIT in Boston, Massachusetts.

Visit their wiki
View moreIGEM Uppsala 2012
 The team of Uppsala iGEM 2012
The team of Uppsala iGEM 2012The project of 2012's goal was combat the rising threat of antibiotic resistance. The team of 2012 used a method of inhibiting translation of resistance genes by the use of small RNAs. The project also continued to express chromproteins and send these to the registry.
 ... thath's how resistance is futile!
... thath's how resistance is futile!
A part of the initial procedure was to screen for sRNAs. This was done by the use of a screening procedure where random oligonucleotides were inserted in the sRNA framework. The device expresses a response when a sRNA is transcribed which inhibits a chosen target sequence, in this case the resistance gene. This sRNA (coupled with a stabilizing spot42 scaffold) was produced and was tested for inhibition of translation of resistance proteins.
Human Practice
The main part of our human practice was focused on organizing an iGEM Scandinavian meeting. By organizing this day we let different iGEM teams from scandinavia get a chance to present their projects and share their experiences.
Result
In the regional championship in Amsterdam, we received a Gold medal for our work with effectively down regulating antibiotic resistance in a multiresistant strain.

Visit their wiki
View moreIGEM Uppsala 2011
Project Setting the color
 The team of Uppsala igem 2011
The team of Uppsala igem 2011The iGEM project 2011 focused on regulation of gene expression through the help of light. This was also the first year that the very promising chromoproteins were first introduced in iGEM Uppsala. The project was based on constructing a system capable of detecting multiple wavelengths. Most systems focused on a one light regulated systems although iGEM 2011 wanted to take it one step further and build a system that could be regulated by multiple different wavelengths. The system was designed to regulate expression of three different genes independently depending on what wavelength that was used. The goal in the end was to prove the concept by growing a colorful picture of bacterial culture, much like Andy Ellington's coloroids picture.
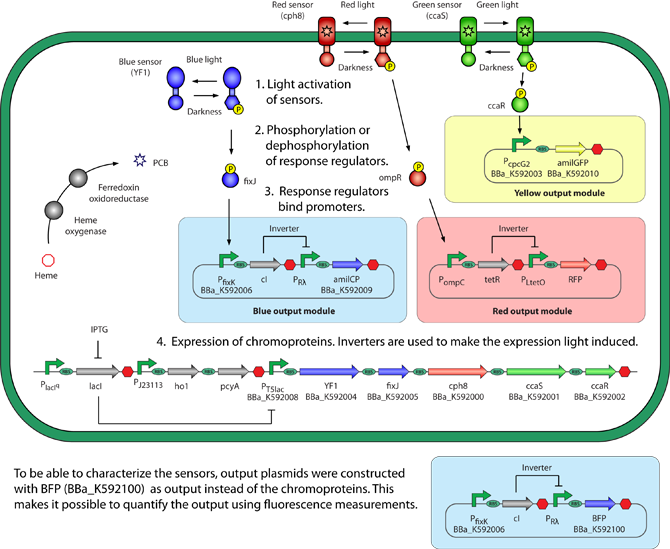 The figure describes the approach in four discrete steps. In step 1 the sensors are activated, step 2 phosphorylation or desphosphorylation of response regulators, step 3 binding of response regulators to promoters, finally in step 4 the expected color (chromoprotein) is expressed
The figure describes the approach in four discrete steps. In step 1 the sensors are activated, step 2 phosphorylation or desphosphorylation of response regulators, step 3 binding of response regulators to promoters, finally in step 4 the expected color (chromoprotein) is expressed
Human Practice
The human practice project of iGEM Uppsala 2011 was based on reaching out and making people aware of the many possibilities of Synthetic biology and GMO. This was done by simultaneously reaching out to media and also to high schools in order to spread the possibilities of synthetic biology. The team were featured on various journals such as “Biologen” and they also held presentations in various places were one of these was the swedish high school “Katedralskolan”.
Result
The team managed to get parts of their system working and because of this they managed to go forward to the finals in MIT (Massachusetts institute of technology) in Boston after have won a gold medal in the regional championships in Amsterdam.
Visit their wiki
View moreIGEM Uppsala 2010
"Levande Klocka" (Living clock)
 The team of Uppsala iGEM 2010
The team of Uppsala iGEM 2010The iGEM Uppsala project of 2010 “Living Clock” involved building a concentration band detection sensor in order to create a bacterial clock. To prove the concept further the the plan was to demonstrate its usefulness by building a bio-clock and a color display unit.
The goal was to produce a concentration band component that had the ability to quantify a signal. The component itself was to be able to be combined with chemical sensors in order to enhance the signal. By using the concentration band in combination with regulated transcription factors one could make a color display unit. Each pixel of the Color Display Cell (CDC) consists of three colors that can be controlled individually. Through full control of these pixels one could produce complex color pictures and possibly even movies if combined with an efficient color degrading system controlled by an electrical current.
Results
The iGEM team of Uppsala managed to get a bronze medal in the championships in MIT (Massachusetts institute of technology) in Boston.
Visit their wiki
View moreIGEM Uppsala 2009
Sun to Alcohol: Booze Bugs

The iGEM Uppsala team of 2009 focused on alternative attempt to harvest energy with the sun as the primary energy source. Crude oil resources are getting more and more depleted in todays society and more importantly the effects on the climate demands a shift of how energy is produced today, in order to do that it is imperative to find a more sustainable energy source compared to the ones that exist today. The goal of the project 2009 was to produce biofuel in the form av ethanol and butanol with the use of cyanobacteria, Synechocystis sp PCC6803. Through utilizing the cyanobacterias ability to directly convert sunlight, the iGEM team made engineered constructs for ethanol and butanol production as well as observing strategies to increase the yields of the developed method.
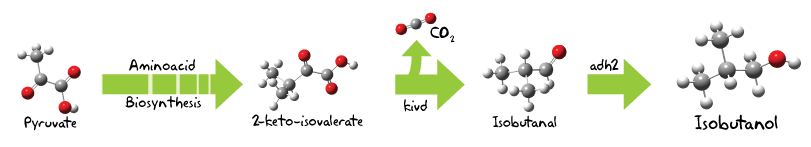
Results
The iGEM team received a bronze medal in the championships in MIT (Massachusetts institute of technology) in Boston.